While involved in various efforts to bring new data into the CDM, I’m noticing a recurring issue that the source data is more specific than the corresponding standard concept. My understanding is that the typical way to resolve this is to expand the standard vocabularies, though I’m starting to think there may be a better fit in certain situations.
Due to the scope of the vocabularies (SNOMED in particular), the information can be adequately represented by referencing a combination of standard concepts. The CDM allows one to many relationships between concepts when a source concept maps to both an observation_concept_id and value_as_concept_id. (Relevant discussion here).
Excerpt from wiki/OBSERVATION:
‘Note that the value of value_as_concept_id may be provided through mapping from a source Concept which contains the content of the Observation. In those situations, the CONCEPT_RELATIONSHIP table in addition to the “Maps to” record contains a second record with the relationship_id set to “Maps to value”. For example, ICD9CM V17.5 concept_id 44828510 “Family history of asthma” has a “Maps to” relationship to 4167217 “Family history of clinical finding” as well as a “Maps to value” record to 317009 “Asthma”.’
Where the relationship conceptually looks like :

In other words, if the concept you are mapping to lacks the specificity regarding the VALUE, then we can map to an additional concept that populates another field. What if, however, the information you are losing in mapping to a more general concept doesn’t pertain to the value of the observation but instead the observation itself?
Building off the previous example, say our source concept is ‘Paternal history of asthma’ and we also want to specify the relationship of the family member. We could do so by creating a new relationship type of ‘Maps to qualifier’ and use it like so:
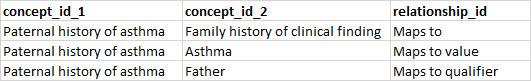
In terms of interoperability, expanding the standard vocabularies to incorporate new concepts as necessary makes sense. However, I’m seeing more and more situations where leveraging an optional data field makes concept mapping more accessible.
For example, at TMC we’re looking at bringing in ECG data and a colleague has developed a source vocabulary that is more detailed than the ECG concepts provided by LOINC, specifically qualifying the wave and direction. Given that the source vocabulary has 10x the number of concepts, which at times are quite specific, extending LOINC would be an endeavor. Though, given the overlap, if the additional details could be specified in another concept, no expansion would be needed.
e.g.

(The above example would require that qualifier, or something similar, be added to the measurement table)
This design has particular relevance to the GIS work group as most of what we are trying to bring into the CDM does not exist within a single concept in the vocabularies but can be adequately represented by a combination of SNOMED concepts.
