nice framing of the problem.
a quick addition in terms of options to explore:
when a researcher manually selects the set of ~20 variables to put into
their table 1, there is presumably some logic behind it. some variables
are likely fixed for all studies because they are universally expected
(age and gender), but the rest are chosen for a particular reason. id be
helpful to dig into what those reasons are. a few possibilities, from my
own experience:
-
the covariate is an expected confounder, so you want to show youve
thought of it. the covariate is either suspected to be strongly associated
with treatment choice, strongly associated with outcome incidence, or at
least a little of both. -
the covariate is highly prevalent, such as a common comorbidity within
the disease state, and its thought to be valuable to communicate that a
large fraction of the population has this other chacteristic (whether or
not its an effect modifier). -
the covariate may be disproportionately prevalent, relative to general
background rate of the covariate or expected value from other cohorts.
since its an ‘unusual’ characteristic of the cohort, even if not highly
prevalent, it may be interesting. clinical example: secondary malignancy
rate prior to exposure in a cohort of new users of some chemotherapy. -
the covariate was notably imbalanced prior to statistical adjustment
(e.g. propensity score matching in comparative cohort analysis), so you
want to show why groups may have not been comparable. -
the covariate is notably imbalanced after statistical adjustment, and
you want to show one of the potential sources of residual bias.
im sure there are other reasons, and id be great to use this thread to
capture them here.
for each of the reasons, it seems it should be possible to define a
heuristic that that could be applied to the ‘large scale characterization’
results we currently generate, to pick out the interesting variables and
present them in a simple form. i would strongly argue that this should be
done in addition to, not instead of, sharing the full large scale
characterization results, but the small table 1 can make its way into the
main body of a manuscript and the larger results likely require an
interactive dissemination strategy that would be considered supplementary.

 ). However, as far as I can see STROBE is not very specific on what should go into table 1. Here’s what I could find for cohort studies:
). However, as far as I can see STROBE is not very specific on what should go into table 1. Here’s what I could find for cohort studies: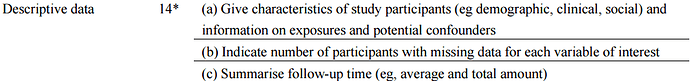
 You only hear the few who complain. The ocean of happy followers and Martijn admirers - are silent.
You only hear the few who complain. The ocean of happy followers and Martijn admirers - are silent.