Hello Everyone,
I came across the T2DM Phenotype algorithm in PheKB website click here and have few questions.
I am a graduate trying to learn what are phenotypes and how it works etc. Not from healthcare background. So would really appreciate if domain jargons are minimized
Can you help me with the below questions?
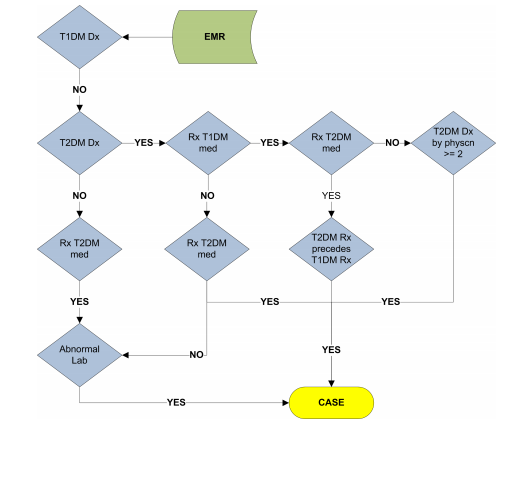
Let’s breakdown the flowchart (pasted below) into individual paths
path 1) EMR -----> T1DM DX -----no–> T2DM DX ----no—> RX T2DM MED —yes—> Abnormal Lab —yes----> CASE
Q1) - How can the patient be prescribed for T2DM meds when he is not diagnosed for T2DM?
path 2) EMR -->T1DM DX —no—> T2DM DX ----yes—> RX T1DM MED —no—> RX T2DM MED —no----> Abnormal Lab —yes----> CASE
Q1) Based on path 1 and path 2 what I can infer is, if the patient has abnormal lab, he is a T2DM patient irrespective of whether he was prescribed or not for RX T2DM meds
path 3) EMR ----->T1DM DX —no----> T2DM DX —yes—> RX T1DM MED —yes—> RX T2DM MED ----yes—> T2DM RX precedes T1DM RX —yes—> CASE
Q1) Again, how can a patient be prescribed with T1DM meds when is not diagnosed for T1DM ?
Q2) It makes sense to see T2DM meds as patient was diagnosed for T2DM but why do we see it as T2DM RX precedes T1DM RX mean? Shouldn’t it be T1DM RX precedes T2DM RX? Because when I did a google search, I found out that T1DM` usually occurs before once enters adulthood. So Am I misinterpreting this?
path 4) EMR ----->T1DM DX —no----> T2DM DX —yes—> RX T1DM MED —yes—> RX T2DM MED ----no—> T2DM DX by Physician >=2 —yes—> CASE
Q1) In this path, a patient is diagnosed for T2DM but he has been prescribed T1DM meds. Is this possible and when can this happen?
Q2) Similarly, though the patient is diagnosed for T2DM he isn’t prescribed for T2DM Meds. Is it like patients are prescribed meds only after multiple visits or something?
Can you help me with the above questions please? Will really be helpful
Thanks
Akshay

