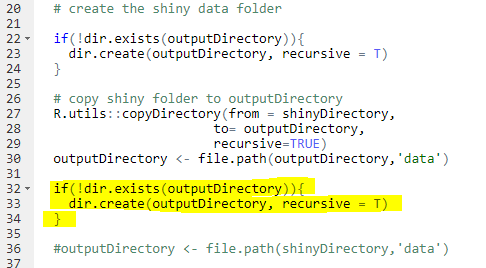
Hi all,
I developed a Targeted and Outcome cohort using Atlas. I followed the instructions to customize the R-package but I receive every time an error while I can successfully build the package. For example, in the CodeToRun.R function I receive an error that there is no file called “setting.csv” if I want the argument createShiny = T in the function execute. If I leave createShiny = F and follow the instructions in the readme.md file I receive the error that the connection (org.postgresql.util.PSQLException) failed. However, I can access the database with my credentials in Jupyter Notebook.
I feel like I am missing something and I hope someone can help me.
Thank you in advance!
Sebastiaan
BTW, this is specified in my logs.txt
| 2021-03-02 10:16:08 | [Main thread] | INFO | ERSPCdiagnosisdeath | execute | Creating cohorts |
|---|---|---|---|---|---|
| 2021-03-02 10:16:08 | [Main thread] | FATAL | rJava | .jcheck | Error in rJava::.jcall(jdbcDriver, “Ljava/sql/Connection;”, “connect”, : org.postgresql.util.PSQLException: The connection attempt failed. |
| 2021-03-02 10:19:41 | [Main thread] | WARN | utils | read.table | cannot open file ‘./ERSPCdiagnosisdeathResults/settings.csv’: No such file or directory |
| 2021-03-02 10:19:41 | [Main thread] | WARN | utils | read.table | cannot open file ‘./ERSPCdiagnosisdeathResults/settings.csv’: No such file or directory |
| 2021-03-02 10:19:41 | [Main thread] | FATAL | utils | read.table | Error in file(file, “rt”) : cannot open the connection |
| 2021-03-02 10:19:41 | [Main thread] | FATAL | utils | read.table | Error in file(file, “rt”) : cannot open the connection |
| 2021-03-02 10:21:45 | [Main thread] | INFO | ERSPCdiagnosisdeath | execute | Creating cohorts |
| 2021-03-02 10:21:45 | [Main thread] | INFO | ERSPCdiagnosisdeath | execute | Creating cohorts |
| 2021-03-02 10:21:45 | [Main thread] | INFO | ERSPCdiagnosisdeath | execute | Creating cohorts |
| 2021-03-02 10:21:45 | [Main thread] | FATAL | rJava | .jcheck | Error in rJava::.jcall(jdbcDriver, “Ljava/sql/Connection;”, “connect”, : org.postgresql.util.PSQLException: The connection attempt failed. |
| 2021-03-02 10:21:45 | [Main thread] | FATAL | rJava | .jcheck | Error in rJava::.jcall(jdbcDriver, “Ljava/sql/Connection;”, “connect”, : org.postgresql.util.PSQLException: The connection attempt failed. |
| 2021-03-02 10:21:45 | [Main thread] | FATAL | rJava | .jcheck | Error in rJava::.jcall(jdbcDriver, “Ljava/sql/Connection;”, “connect”, : org.postgresql.util.PSQLException: The connection attempt failed. |