Another possible source of metadata examples are those utilized by Arachne catalog/framework from Odysseus. Taging @Dymshyts to perhaps talk him into reply with metadata they are considering?
Vojtech - looked at how the proposal is shaping up and it looks fantastic, great progress. Here are my thoughts on this topic.
To enable seamless data and analysis design exchanges, including across multiple models, technologies and implementations, there are two models that need metadata - Data Model (OMOP CDM) and Business Process Model (Observational Study Model). I will comment on both.
The OMOP CDM data model is well described today, except for the metadata for the model and data set itself. which is what we are trying to tackle in this group
Arachne is heavily relying on the use of meta data for both process and data modes.
Here are some some additional possible metadata describing CDM model:
- ModelType: {OMOP, I2B2, Sentinel etc…}
- ModelVersion: {4, 5, 6 etc…}
- Revision: {1,2,3,4…}
- Organization: OHDSI
Some possible metadata for the Data Set:
- Data Type Included: {Claims, EHR, Survey, Registry, Lab, ClinicalTrial…}
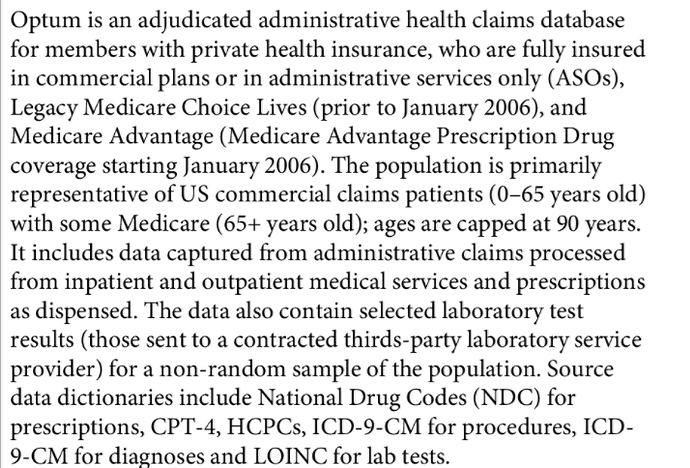
- Vendor: QuintilesIMS, Truven, Optum, CPRD, CMS etc…
- Version (optional)
- Vocabulary Used: {}
- Vocabulary Version
- Last Updated Date
- Year Start
- Year End
An important point to add is that Arachne Data Catalog is also taking an advantage of Achilles generated stats to describe registred data sets.
Another very useful thing for us could be is to describe and agree on a common Business Process Model view, start with a simple conceptual model. Here are a few important examples:
- Study
- Cohort
- Concept and Concept Set
- Analysis
- Analysis Data Results
We should think about a set of common metadata elements for those…
- Guid
- Name
- Description
…as well as element specific, deep, design related meta data, which ATLAS already is doing a good job generating today in their JSON
Sorry for the delay.
My typical metadata would be:
- The data table is sampled randomly from the population
- The data table is biased towards XYZ (working age population with commercial insurances, men as former military servicemen, procedures that billibable)
- The data table is completely capturing the patient
- The data table is sampling from patients
- The drug table doesn’t contain immunizations
Etc. All of these will help the users to assess, whether the denominator or numerator of a score can be trusted or not.
We will have different OMOP instances for our different data sources. All will have their little nuances. We are still working on our first source to OMOP ETL, so I am sure we will find more data for the metadata table. Some examples I have come across:
- The procedures table does not contain surgery data
- The provider specialty field isn’t completely accurate/updated by the source
- The drug table includes Ordered medications which were mapped to the Clinical Drug concept class
- The death table data comes from EHR and public health sources
Thank you for the reply so far. I will prod the remaining players. @rimma @Ajit_Londhe.
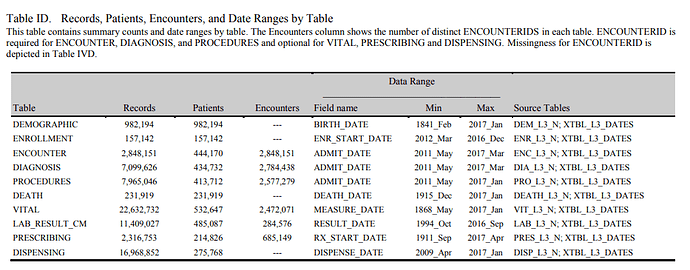
The data below can be obtained from Achilles pre-computations. I like to focus on metadata that can not be inferred from Achilles. I would like to define the scope of metadata and information that is “outside Achilles reach”.
The procedures table does not contain surgery data
The drug table doesn’t contain immunizations
Here are some examples of metadata/provenance that we found important at the NYC-CDRN project and Montefiore.
- In multi-institutional integrated OMOP instance, identification of the source (hospital) for each record to support by hospital/comparative analysis and data filtering.
- Data completeness that cannot be assessed from the data. For example, death data are complete and sourced from death index up to 2011. Since 2012, they are derived from two sources, in-hospital recorded deaths and follow-up.
- Attribute definition/derivation. For example, visit start date/time for inpatient admissions through ER may represent a different time stamp at different hospitals: triage timestamp at one and room placement at the other.
- Date/time precision may differ even between the data coming from different sources at the same hospital. Critical for analysis.
- Other examples
- Many procedure records are duplicated because data are sourced from two systems.
- Measurements are not explicitly linked to visits in the source. Linkage can only be inferred by dates.
- Blood pressure measurements are not always paired (systolic and diastolic) in the source.
Please note, that granularity of the metadata varies. It can be a whole data set, a data subset, a domain/table, a domain attribute, a record.
Metadata proposal in the new GitHub based infrustructure. https://github.com/OHDSI/CommonDataModel/issues/79
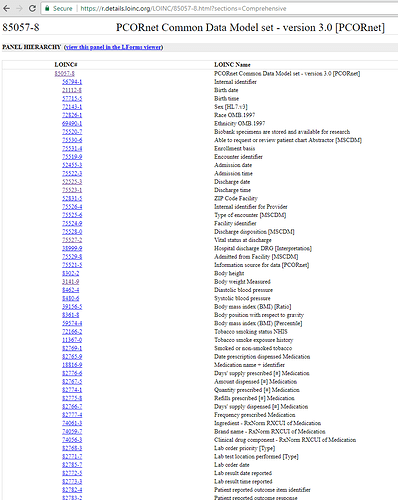
Use Case: We have been working on ETL’ing into PCORnet CDM by way of OMOP. That is we ETL from source to OMOP and then from OMOP to PCORnet. Included is an example of metadata that PCORnet wants in their CDM, that could be stored in OMOP with the proposed Metadata table.
Metadata:
What is the data management strategy currently present in the ENR_START_DATE field on the ENROLLMENT table. ( The PCORnet enrollment is similar to OMOP Observation_period). The possible values are:
- 01=No imputation or obfuscation
- 02=Imputation for incomplete dates;
- 03=Date obfuscation;
- 04=Both imputation and obfuscation;
Nice. I like that. Question: Is this metadata or is this an additional field in the OBSERVATION_PERIOD?
In PCORnet it is a series of explicit columns with questions like the
above. But obviously could be handle using the metadata table and set of
conventions for the metadata name since there will not be a standard
concept. My point was to provide an example of how we might use the
metadata table. And I am NOT advocating OMOP adopt any structure based
upon the PCORnet CDM.
This is another call to comment on the metadata proposal if you have data that would not fit the proposed structure.
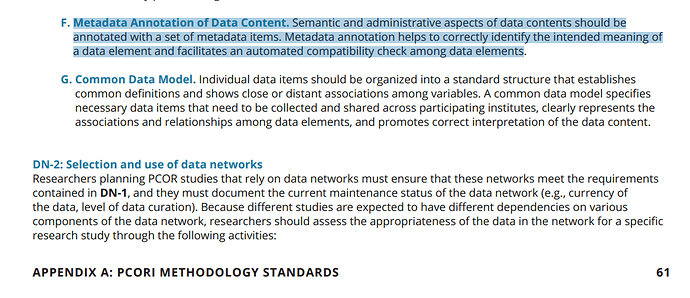
Additional push for getting this table in is point F under DN-1 requirement for data networks in this PCORI report. http://www.pcori.org/sites/default/files/PCORI-Methodology-Report.pdf
(go to page 61)
pasted excerpt below
The metadata table is now part of CDM.
In phase 2 - we hope to shape the conventions on concepts and use of it.
I was asked to comment on the question asked elsewhere (and pasted below)
Hi CDM Builders!
I am trying to populate cdm METADATA table and would like to ask you a couple of questions:
- I see that in the proposal METADATA_CONCEPT_ID populated with non-standard concepts. Can it be populated with non-standard concepts?
- What are the possible values for METADATA_TYPE_CONCEPT_ID field?
- How to populate VALUE_AS_CONCEPT_ID field?
Thank you in advance!
-
If the concept is not present - use 0. If you have local concept (2B+numeric space) - you can use that
-
@Ajit_Londhe would be best to answer that. There was some discussion about it in the issue tracker on github
-
As done in measurement or observation table. For example if value for type of data = clinical trial, you use value_as_concept_id of Athena
Ad concepts for each table in CDM - do we have a convention for that @Patrick_Ryan?
For example PCORNet folks managed to talk LOINC into creating even LOINC codes for their CDM
Another example relevant to metadata is standardizing database name and database description.
Example description can be found in this article
https://doi.org/10.1371/journal.pone.0192033.s018
In Table 1 Inferring pregnancy episodes and outcomes within a network of observational databases
Pasted here as picture
Database name is already captured in CDM_SOURCE table.
Another example of metadata (and public “Achilles-like” US data)
(we should learn from our PCORnet colleagues)
http://www.gpcnetwork.org/?q=DataCharacterization
table from this report
http://www.gpcnetwork.org/sites/default/files/pictures/C4UN_20170412_EDCRPT-trimmed.pdf
(a total of 5+ sites is presented in the first link)
Hi @Vojtech_Huser ,
Have we (as a community) been using metadata tables very frequently in network studies? I didn’t notice discussion of the metadata tables in the Save Our Sysephus challenge, but i may have missed it. @clairblacketer , hope you don’t mind me copying you, but just wondering if there has been discussion about evaluating site level metadata, as stored in the metadata table, during the data diagnostics step or the interpretation step of the network study pipeline?
Some of the above limitations of a dataset, as mentioned by folks in this conversation, could have implications for how much we trust data from a particular site.
Thank you for your contributions, gang!