Hi all,
Apologies if this has been done in another topic (if so I couldn’t find it) but we’re interested in how to model some of our NLP-derived facts in NOTE_NLP - specifically, NLP-derived numeric values.
For some of our NLP work, the mapping is relatively straightforward. For example, we have an existing pipeline to extract ICD-9 and ICD-10 codes from pathology reports - here, we can just map the CONCEPT_ID for the ICD-9/10 code to note_nlp_source_concept_id and then use CONCEPT_RELATIONSHIP to determine the standard code, and insert that as note_nlp_concept_id.
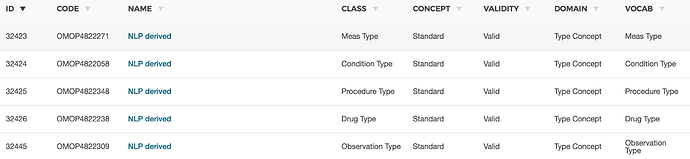
However, some of our NLP pipelines extract numeric values from freetext - for example, we have one in place that extracts numeric PHQ-9 values, and one that extracts LVEF from echocardiogram reports. There don’t really seem to be any concepts, standard or otherwise, for these continuous variables, e.g. “PHQ-9 value of 15.”
Our initial plan was to use a CONCEPT_ID corresponding to the measurement as the note_nlp_concept_id, then simply insert the extracted value as lexical_variant. However, I’m curious to hear how others have approached this issue - I’m sure we’re not the first to encounter it.