If you told me there was a disease that occurs in 10% of patients at risk, which is one of the leading causes of infertility and a source of substantial decreases in quality of life, then I’d expect that already know just about everything we need to know about the disease natural history and the effects of available treatments. But it turns out that, despite the significant public health impact of endometriosis to women of child-bearing age, there is still much we need to learn and lots of opportunities to contribute.
Now, I have to confess that before a couple years ago, I’m pretty sure I couldn’t have told you what endometriosis was. One of the most satisfying aspects of being part of the OHDSI community for me is getting the opportunity to meet and collaborate with others who I can learn from. It turns out our own @noemie is not just a world expert in machine learning, but also an active researcher in endometriosis and sits on the Endometriosis Foundation of America’s Scientific Advisory Board. So when she raised my awareness about the disease, I became very excited by all the possibilities about how evidence from observational data could be legitimately useful to researchers, providers and patients by providing a real-world perspective about the disease natural history. If you want to be similarly inspired, read or watch @noemie’s talk from 2016 at Patient Awareness Day. Now @noemie’s message was about “Giving patients the tools to contribute to endometriosis research”. I’d like to augment that by “giving patients the evidence from endometriosis research” and I think the OHDSI community is well-positioned to deliver on that.
The question is: where can we start? It turns out the endometriosis research community has already done us a great service: just last year, they published a consensus statement in Reproductive Services, “Research Priorities for Endometriosis: Recommendations From a Global Consortium of Investigators in Endometriosis”. In the publication, they outline several areas of opportunity that seem right up our community’s alley for anyone who may be interested.
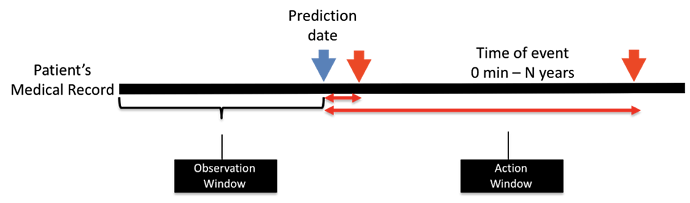
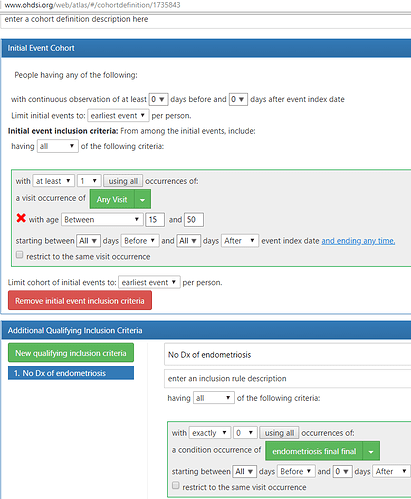
One particular topic that @noemie and I would like to explore this month: among a target population of women of child-bearing age who visit a doctor and have not yet been diagnosed or treated for endometriosis, which patients will go on to be diagnosed with endometriosis in the next year? We plan to use ATLAS to develop a phenotype for endometriosis and characterize the population, and then apply OHDSI’s PatientLevelPrediction R package (thanks @jennareps and @Rijnbeek!) to determine how well we can correctly identify patients who will eventually develop the disease using baseline characteristics on or prior to the index visit. And if we get a predictive model that shows promise with high discrimination, good calibration, and holds up over internal and external validation, we can consider how to make a parsimonious model that could become a risk calculator for use by interested providers and patients. At least that’s our thought going into the project…we’ve got a month to sort out the details and see how it all plays out.