I verified that my prereqs worked for my install by testing at the following link: https://ohdsi.github.io/Hades/rSetup.html#Verifying_the_installation
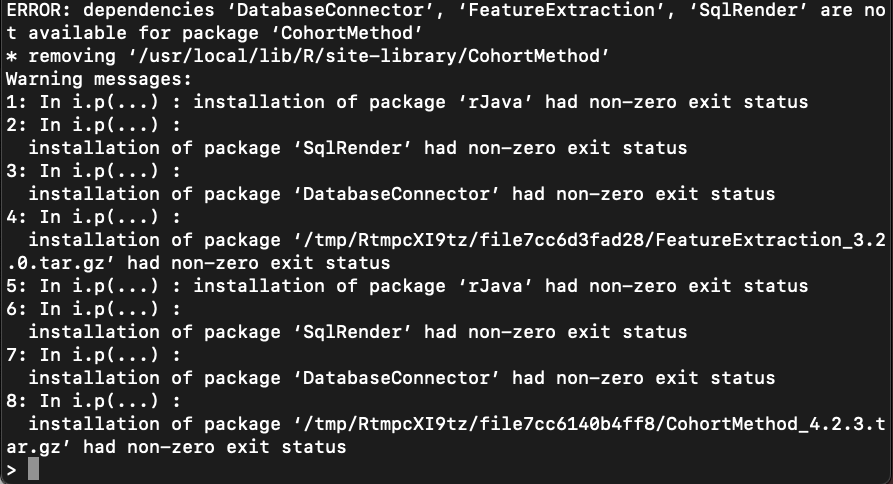
The first command works fine but when I run I get an error on install. I have tried to install the cachem in Rstudio without any luck. This may be an issue with Rstudio but I am getting the issue when I follow the install insturctions at
https://ohdsi.github.io/Hades/installingHades.htmlinstall.packages(“remotes”)
remotes::install_github(“OHDSI/CohortMethod”) as well.
- installing source package ‘cachem’ …
** package ‘cachem’ successfully unpacked and MD5 sums checked
** using staged installation
Warning in as.POSIXlt.POSIXct(x, tz) : unknown timezone ‘GMT’
** libs
Warning: R include directory is empty – perhaps need to install R-devel.rpm or similar
*** arch - i386
"C:/rtools40/mingw32/bin/“gcc -I"C:/PROGRA~1/R/R-40~1.3/include” -DNDEBUG -O2 -Wall -std=gnu99 -mfpmath=sse -msse2 -mstackrealign -c cache.c -o cache.o
cache.c:1:10: fatal error: R.h: No such file or directory
#include <R.h>
^~~~~
compilation terminated.
make: *** [C:/PROGRA~1/R/R-40~1.3/etc/i386/Makeconf:222: cache.o] Error 1
ERROR: compilation failed for package ‘cachem’