I will continue within this thread because it was the start of hours of thoughts on how to map ECLS data. It seems that neither SNOMED CT nor any other terminology can provide a convenient and sufficient way because they lack the respective concepts.
One major player in regard to ECLS data and databases is ELSO. ELSO for the ones that have never heard it is the Extracorporeal Life Support Organisation which was founded by the inventor of ECMO itself, Dr Bartlett. It has strict requirements of what to map. Based on their data base structure we have come up with one possible solution.
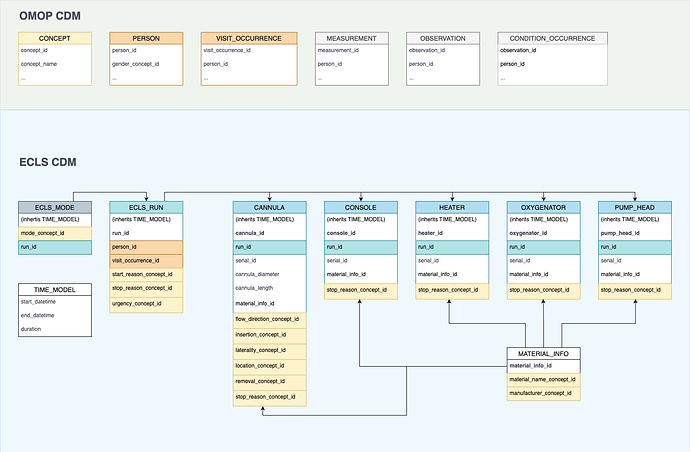
I would like to propose the idea of the extension ECLS-CDM similar to the Genomic-CDM that already helps map genome data in an OMOP-compliant fashion.
The goal was to map as much data as possible in a generic OMOP fashion in the respective tables (such as PERSON, MEASUREMENT, VISIT_OCCURRENCE, …) while creating some additional tables that behave less generic and store data that specific to ECLS therapy. Influenced by ELSO we tried to keep the naming similar.
- ECLS_RUN: It is the singular connector to the OMOP CDM and contains additional information as to why ECLS therapy was initiated or stopped and if it was an emergency procedure or elective.
- ECLS_MODE: will store information about the mode of operation (VV, VA, VVA, PECLA, etc.)
- CANNULA: contains information about the cannulas in place, their flow direction and where they have been placed
- CONSOLE: contains information about the console in place (e.g. CardioHelp)
- HEATER: contains information about the heating unit if there was one installed (e.g. Maquet HU35)
- OXYGENATOR: contains information about the oxygenator
- PUMP_HEAD: contains information about the pump head (ECLS consoles can be combined with different oxygenators and pump head depending on the local requirements)
- MATERIAL_INFO: stores manufacturers and product names (e.g. Getinge Avalon)
- TIME_MODEL: contains fields that the other tables will inherit. This is because the tables were created with Django and Python’s class inheritance feature.
Is this something that OMOP CDM is interested in supporting officially?
More work is necessary to create a custom ECLS vocabulary that contains the respective concepts necessary to meet ELSO criteria as well as easy mapping of ECLS data.
We have been in contact with EuroELSO (the EU brand of ELSO) and it seems like they are interested in an open-source, easy-to-share, improvable CDM that will be maintained by the community.
This is of course preliminary work and can be subject to major changes. With this in mind I thank you for your thoughts on that topic.