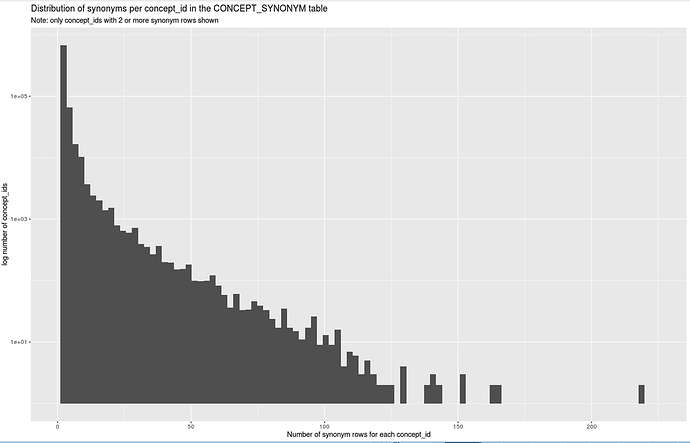
Just delving into the CONCEPT_SYNONYM table a bit further to determine whether we can use it as part of the source-to-OMOP concept mapping system in our ETL processing…
So there are a lot of concept_ids which have large numbers of rows in the CONCEPT_SYNONYM table. The question is how many of these are currently erroneous?
To answer that, I did a series of spot checks on concept_ids with 50 or greater synonym rows in the CONCEPT_SYNONYM table. My conclusion is that the vast majority of those are fine and all the rows are true synonyms, albeit with a lot of redundancy, but that’s OK. None of the affected concepts are standard concepts, afaics. A few seem to include synonyms of descendants, which is not strictly desirable. But very few are outright incorrect, as with the examples given in the OP.
Given all this, we’ll proceed with using the CONCEPT_SYNONYM table in our ETL mapping code, but will check the next vocabulary release to see if erroneous synonyms have been removed.