Hello,
I am running into this same error. Has a solution been found? I can create a cohort definition but it fails when try to generate. Using latest Broadsea docker application against a fresh CDM 5 repository on PostgreSQL.
Thank you
Rick
Your best bet is to go into the Atlas UI, go to the Export->Sql tab, and copy the postgreSQL statements and run them individually. The entire cohort SQL was stored in the error field, but the error field is not large enough to contain the entire SQL so it is truncated.
Let us know if you narrow down which part of the query is giving you the error. Remember, run them one statement at a time so you can get to the specific statement that gives you the error. If you run it as a batch, it may not give you the detail you need.
The first statement causes an error
CREATE TABLE #Codesets (
codeset_id int NOT NULL,
concept_id bigint NOT NULL
)
;
ERROR: syntax error at or near “#”
SQL state: 42601
Character: 14
That’s the MSSQL rendering, under the SQL tab, there’s dialects, pick the PostgreSQL one, and use that.
Sorry. Changed the dialect and it is the insert statement
INSERT INTO Codesets (codeset_id, concept_id)
SELECT 0 as codeset_id, c.concept_id FROM (select distinct I.concept_id FROM
(
select concept_id from @vocabulary_database_schema.CONCEPT where concept_id in (1503297)and invalid_reason is null
) I
) C;
From the Broadsea docker-compose.yml
datasource.cdm.schema=cdm
datasource.ohdsi.schema=ohdsi
flyway_schemas=ohdsi
flyway.placeholders.ohdsiSchema=ohdsi
Did I miss a location to define the vocabulary_database_schema?
The replacement tokens comes from the ohdsi.source_daimon table. The @vocabulary_database_schema will use the cdm’s schema in the event that the soruce_daimon table doesn’t declare a Vocabulary daimon table qualifier.
Long story short, you can ignore all the settings in docker-compose, just replace @cdm_database_schema with your cdm schema, and @vocabulary_database_schema also with your cdm schema (unless you have an environment where you put your vocabulary tables in a separate schema. I’m assuming you did not since this is coming from Broadsea.
Then run the script and try to determine where the failure is.
-Chris
Everything runs fine down to the delete statement. What should I use for the 3 target tokens?
@target_database_schema
@target_cohort_table
@target_cohort_id
Side note - While running the statements, I also verified that my source daimon table contains the correct schema values.
@target_database_schema = your results schema
@target_cohort_table = cohort
@target_cohort_id = your cohort definition_id (which you can see in the URL of atlas when you are editing the cohort definition.
If all this works out,then the issue could be in the ‘inclusion analysis statistics’ part of the query, which is not visible via the SQL export (since they do not contribute to constructing the cohort, but they are merely build-time statistics that we capture and save to the results schema.
Are you sure that you have the cohort_inclusion_* tables created in your results schema? You can find this schema by going to your WebAPI endpoint to: WebAPI/ddl/results?dialect=postgresql
Also it would help to know what version of Atlas and WebAPI your instance of Broadsea has. If this turns out to be missing tables, we may need to ask @lee_evans to update the broadsea instances to include the correct results schema with the bundled CDMs.
The delete statement failed due to a missing final_cohort table.
DELETE FROM ohdsi.cohort where cohort_definition_id = 4;
INSERT INTO ohdsi.cohort (cohort_definition_id, subject_id, cohort_start_date, cohort_end_date)
select 4 as cohort_definition_id, person_id, start_date, end_date
FROM final_cohort CO
;
I have 3 cohort_inclusion tables - cohort_inclusion, cohort_inclusion_result, cohort_inclusion_stats.
The Atlas home page states that I am using version 2.6.0 of Atlas and WebAPI.
@Chris_Knoll FYI Broadsea doesn’t currently bundle a (postgres) database results schema or any CDM database tables. It deploys the latest released ATLAS/WebAPI (currently v2.6.0).
Ah, so, @rkiefer, what you’ll need to do is create a new schema in your CDM database, and use the WebAPI endpiont: WebAPI/ddl/results?dialect=postgresql to get the DDL. You use this DDL to create the results schema tables. Note: there are a few table initialization scripts that will run forever if you do not have the proper indexes on your tables, please see this discussion for details:
We had run the results ddl previously and there are 82 tables in the schema. When I search the results ddl for postgresql, there is no create statement for the final_cohort table. What is the normal number of tables which should be created?
final_cohort table is a temp table in the generation script, which means it does not get created as a permanent table in the results schema.
The normal number of tables in the results schema are the tables that come out of the /ddl/results endpoint. It’s around 30 (not 81). Here is the list from my environment, note this may have a couple of extra tables compared to the version of the code that you are running (since the latest master has a few new results tables compared to version 2.6):
| TABLE_SCHEMA | TABLE_NAME |
|---|---|
| cknoll1 | ACHILLES_analysis |
| cknoll1 | ACHILLES_HEEL_results |
| cknoll1 | ACHILLES_results |
| cknoll1 | ACHILLES_results_derived |
| cknoll1 | ACHILLES_results_dist |
| cknoll1 | cc_results |
| cknoll1 | cohort |
| cknoll1 | cohort_censor_stats |
| cknoll1 | cohort_features |
| cknoll1 | cohort_features_analysis_ref |
| cknoll1 | cohort_features_dist |
| cknoll1 | cohort_features_ref |
| cknoll1 | cohort_inclusion |
| cknoll1 | cohort_inclusion_result |
| cknoll1 | cohort_inclusion_stats |
| cknoll1 | cohort_summary_stats |
| cknoll1 | concept_hierarchy |
| cknoll1 | feas_study_inclusion_stats |
| cknoll1 | feas_study_index_stats |
| cknoll1 | feas_study_result |
| cknoll1 | heracles_analysis |
| cknoll1 | HERACLES_HEEL_results |
| cknoll1 | heracles_periods |
| cknoll1 | heracles_results |
| cknoll1 | heracles_results_dist |
| cknoll1 | ir_analysis_dist |
| cknoll1 | ir_analysis_result |
| cknoll1 | ir_analysis_strata_stats |
| cknoll1 | ir_strata |
| cknoll1 | nc_results |
| cknoll1 | pathway_analysis_events |
| cknoll1 | pathway_analysis_stats |
In my env, my schema is cknoll1. You should have separate schemas for your CDM tables and your results tables.
Thanks @Chris_Knoll for your help. I worked with @rkiefer some more on this. In the logs of the Broadsea Docker image there was a nested exception which said this part of the query failed:
delete from ohdsi.cohort_inclusion_result where cohort_definition_id = 4 and mode_id = 0
This does not appear in the exported postgres SQL from ATLAS, when we were testing by hand.
The problem is that mode_id didn’t exist in the table. We cross-referenced against the WebAPI endpoint you noted (WebAPI/ddl/results?dialect=postgresql) and saw that mode_id was missing from a few tables. We manually fixed this, and queries are running now. Unsure how this happened, but will backtrack through our setup for ideas.
Right, there is a portion of the cohort generation in Atlas that generates the inclusion rule stats for reporting the inclusion rule impact reports (which is not necessary to do cohort generation, but is informative when looking at the report in Atlas).
Are you saying that the ddl/results script from the end point did not have a mode_id in the table definition? If so, I should make sure that the tables are properly scripted out, otherwise others will have this same problem.
The DDL scripts at the WebAPI endpoint did have the mode_id in the table definition. Sorry for the confusion. That gave us the right DDL so we could recreate the tables. We’re trying to trace back how the tables were initially provisioned as to why they were missing the column.
@lrasmussen @Chris_Knoll - Can the cohort generation fail due to lack of records matching my cohort definition (Say if my cohort entry criteria results in 0 records, can it fail?
For example, I created a concept set with ‘Warfarin’ and that has yielded results.
So when I used select * from codesets, I am able to find records but when I try the same after running “select * from qualified_events”, the result set is empty.
Can the cohort generation fail due to this? Moreover, I have set the cohort entry criteria as shown below
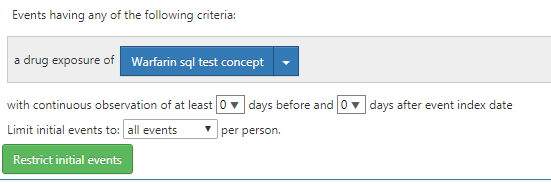
I tried with different settings for ‘iNITIAL EVENTS’ feature. But still am not able to generate the cohort successfully
We have data for Warfarin users. Can you people help us?
I tried running the queries seperately and they all are working fine. But certain queries yields empty table as results but I guess this is due to the criteria specified. Can cohort generation fail due to this?
Thanks
Selva
If you go to the SQL tab under the Export section (within cohort definitions), you can get the SQL that generates the cohort. You can execute it statement-by-statement to understand where you fail to return results.
Hello All,
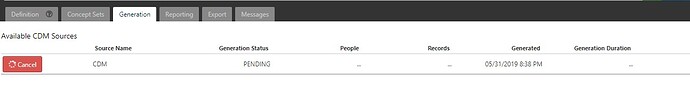
I have set up Atlas at my end and one thing I observed that when I create new cohort and click generate, it says pending and remain stuck at that point.
I clicked Cancel, it doesn’t stop there, so I had to reboot the server.
Once rebooted, I click on Generate Cohort option for the same cohort which was showing pending earlier, runs within 7 seconds.
I am running it against SYNPUF 1k sample data.
Attaching the screenshot.
@Chris_Knoll @anthonysena @Ajit_Londhe (tagging you guys to get some help on this)
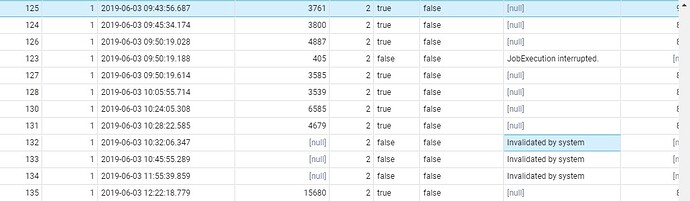
in webapi.cohort_generation_info table, I receive the following message in fail_message column “Invalidated by system”
I am using Postgresql.
TIA
‘Invalidated By System’ messages appears when you restart the service and there’s a job in a ‘running’ or ‘pending’ state. The jobs that were running between the restarts need to be reset (because the thread that is executing the analysis is killed, so on restart, all you can do is mark all running jobs ‘failed’ with ‘Invalidated by system’.
The original problem you were having is strange: you say that you click ‘generate’ but the job just sits in pending status? It’s possible you have another error happening between starting the job and the job starting the execution. Do you have anything in your logs? You mentioned that it did run successfully once. Can you get a simple cohort to generate or revert to the version that was working?